March 2023
A chemical engineering paper suggests supercoiling DNA and gene orientation hold promise for designing precision therapeutics
The genomics revolution has brought the prospect of personalized medicine tantalizingly close. But researchers have yet to achieve a high-resolution picture of the machinery by which genes determine cell form and function. Without this granular understanding, many precision diagnostics and therapies will remain out of reach.
 But now, new studies by Kate E. Galloway, the W. M. Keck Career Development Professor in Biomedical Engineering and Chemical Engineering, and Christopher Johnstone, a fourth-year graduate student in chemical engineering, are filling in important details of this genetic picture.
But now, new studies by Kate E. Galloway, the W. M. Keck Career Development Professor in Biomedical Engineering and Chemical Engineering, and Christopher Johnstone, a fourth-year graduate student in chemical engineering, are filling in important details of this genetic picture.
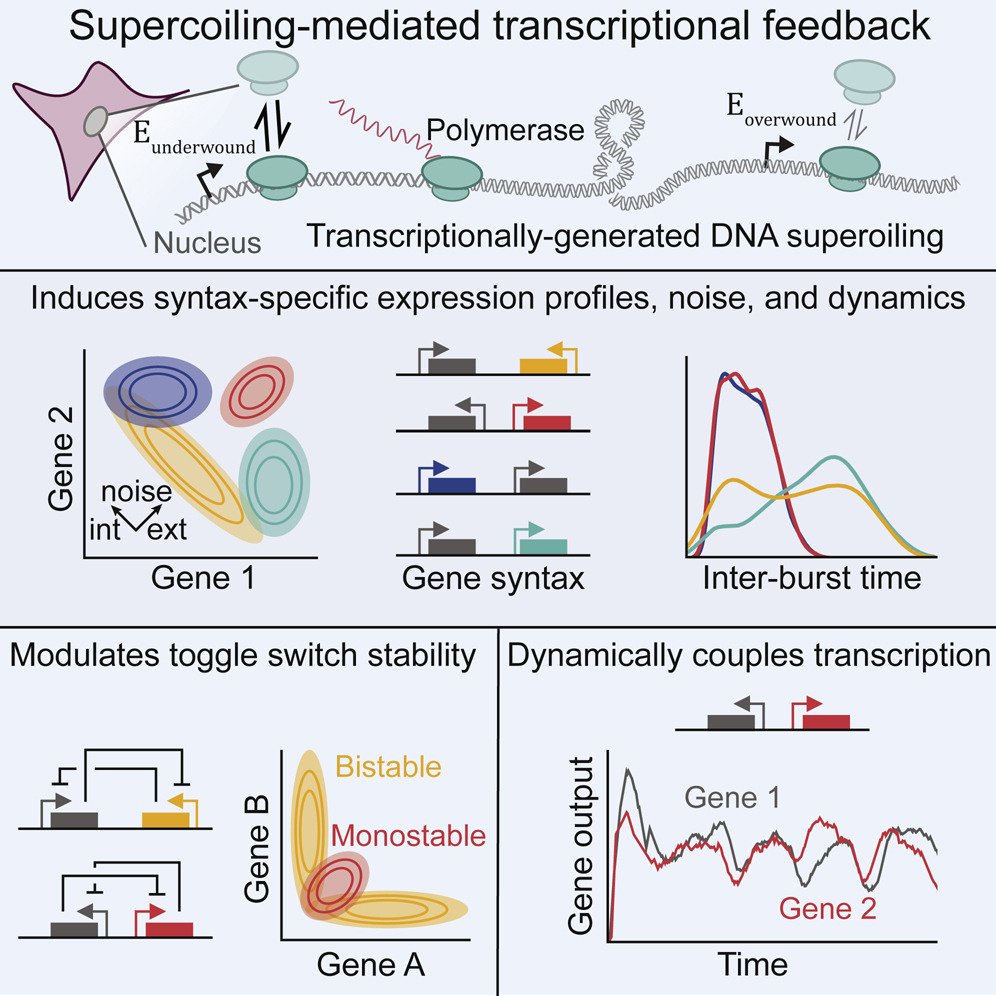
Their focus is supercoiling, a phenomenon in which the DNA double helix molecule winds together very tightly or loosely. This over- or under-twisting of DNA affects how RNA molecules are copied from DNA and create proteins integral to all cellular processes (gene expression), including those that determine whether a cell will develop into skin, kidney or neural tissue.
“We don’t really understand how gene expression works,” says Christopher Johnstone. “Supercoiling is one of those complicated systems designed by nature and we thought investigating it might help us figure out how cells end up doing what they do.”
In their research, Johnstone and Galloway developed detailed computational models of the biophysics underlying certain kinds of gene expression. They demonstrated that the orientation of genes in a region of DNA could determine whether a genetic program central to specific cell functions was switched on or off.
A paper co-authored by Johnstone and Galloway describing this research appears in the October 18 issue of Cell Reports.
 Moving in the right direction
Moving in the right direction
In her synthetic biology lab, Galloway hopes to engineer powerful new approaches for disease modeling, screening for and testing pharmaceuticals and delivering cell-based therapies. To accomplish this task, her lab has been exploring ways of reprogramming cells, to change their identities.
“My group wants to learn how to construct genetic systems that could more precisely control patterns of gene expression in cells,” she says. “Our big-picture vision is to convert commonly-accessible cell types, such as blood cells or skin cells, into cells that are really hard to either access or regenerate, such as neural tissue,” she says. Galloway’s dream is to repair injuries to the brain and spinal cord — an objective that has long challenged medical research.
In her postdoctoral research exploring how to reprogram cell types, Galloway noticed that when two genes known to operate in tandem in the same region of DNA changed position in relation to each other, the outcome of the programming was different.
“She found there was orientation dependence,” says Johnstone, who teamed up with Galloway in January 2020, just months after she arrived at MIT. “If the two genes pointed in the same direction, the correct proteins were expressed, and if they faced into each other, expression failed,” says Johnstone. (A gene starts with a specific sequence of four nucleotides, and RNA polymerase will begin reading in the direction indicated by that sequence. This means, for example, that two genes on the same region of a chromosome that begin with that sequence right after each other are said to be facing the same direction.)
The zebrafish divergence
Why did orientation of such proximal genes matter? Galloway and Johnstone looked to embryo development in zebrafish, a well-documented, real-world gene pair that work in an extremely coordinated way to form the segments of the fish’s spinal column.
“When we investigated closely, we noticed something interesting in our model: when we put these two genes, which are normally on the same piece of DNA in the genome, on opposite pieces of DNA, we lost the coordination ability,” says Galloway.
Like syntax in language — the arrangement of words to make meaningful sentences — the physical relation of genes on a DNA sequence strongly influences the expression of the genes themselves. The correct syntax means that genes encode proteins that will result in proteins forming the building blocks of cell functions.
“This syntax affects the pace with which cells respond to their genetic instructions and whether cells turn certain genes on and others off,” says Galloway. Levels of gene expression matter in the development of cells. But as Johnstone and Galloway found, it is the biophysics underlying this syntax that sets the progression and levels of gene expression in space and over time.
Johnstone began by looking up the gene pair in the zebrafish genome associated with the ordered development of the spinal column. “I figured out how long they are, and their distance from each other, and plugged them into simulations reflecting different orientations of the two genes,” says Johnstone. They wanted to zero in specifically on the impact of supercoiling on the actions of this pair.
“The double helix of the DNA are like two pieces of rope tightly coiled together, and as the RNA polymerase moves through transcribing genetic sequences, it pushes the strands ahead of the RNA to twist more tightly, leaving the strands behind undertwisted and relaxed,” says Galloway.
It’s more difficult for RNA polymerase to bind and move in the direction of overtwisting, and easier for RNA polymerase to slip into undertwisted DNA. If genes are oriented on a region of DNA in a way that encourages undertwisting (facing “away” from each other, or divergently), RNA polymerase meets less resistance, and can slip in more easily to transcribe a genetic sequences. Undertwisting encourages higher levels of gene expression.
If two genes are facing each other or in convergent orientation, RNA polymerase may turn on one gene, then encounter resistance due to overtwisting as it attempts to bind with the related gene. It will leave the second gene “off.” In the case of the zebrafish embryo, they observed, the genes are positioned divergently, so both genes are expressed at the levels required, allowing them to coordinate in producing the proteins that make the spinal cord.
Galloway and Johnstone demonstrated that the orientation of proximal gene pairs profoundly impacts the ability of RNA polymerase to do its job, and hence leads to different levels of gene expression. Supercoiling, they believe, serves as a fundamental, regulatory mechanism. When one gene promotes another to turn on, in the case of divergent genes in zebrafish embryos, there is an explosion in the levels of genes expressed in embryonic cells. “Polymerase loads like crazy, and the two genes start to burst together, becoming coordinated,” says Galloway. “It’s kind of like “Dueling Banjos.’”
Tuning gene expression
Evolution took hundreds of millions of years to produce genetic systems with such precise outcomes. Galloway would like to find some shortcuts. She and Johnstone began developing and testing simple models of genetically programmed gene pairs. One significant result: creation of an A/B switch by means of divergent or convergent gene orientation, which turns messenger RNA (mRNA) on and off. mRNA is a molecule that transfers genetic information from the DNA about protein production to the machinery of the cell itself.
“Although scientists have been building genetic systems with simple on-off gates for a number of years, there has been great variation and a lack of precise control because they have not been accounting for the impacts of physical forces,” says Galloway. “Our modeling actually puts things in a new light, showing that how you compose different systems affects their performance.”
Adds Johnstone, “If you give me some genes and you tell me how they’re oriented and where they are on the DNA, I can tell you something about how supercoiling influences how they’re expressed.” The lab’s models show that it is possible to tune supercoiling by manipulating gene orientation.
Johnstone has successfully translated their computational models in living cells, deploying the divergent and convergent syntax to switch genes controlling fluorescence on and off. Although these studies are still in their early days, their ambitions ride high. “Our models will have an impact on anyone building synthetic gene circuits, whether for gene- and cell-based therapies where more than one gene is expressed or even single genes,” Galloway says.
For Galloway, the potential applications are compelling. “We will make microglia which are useful for modeling Alzheimer’s disease, and we’re going to make neurons to try and repair spinal cords, and we are working on making dendritic cells to go in and fight cancer,” she says. What’s more, the process of generating cells and tissue Galloway is pioneering in her lab is much faster and more precise than current methods, she believes.
“We want to make cells really fast, in seven to 10 days, so we can do disease modeling, drug discovery, and make gene- and cell-based therapies,” she says. “There’s a huge array of things we have applications toward and the only question is how fast we can run at them.”
Funding for this research was provided by the National Institutes of Health.